ToF-SIMS surface bioanalysis

Introduction
ToF-SIMS identifies secondary ions emitted from surface upon bombardment by energetic primary ions (e.g., Bi+, Bi3+ or C60+), usually applied with dose < 1012 ions/cm2 (static regime). It combines surface sensitivity with a great chemical selectivity allowing for discrimination between different biomolecules, based on the differences in their chemical structure. Chemical selectivity is provided by time-of-flight mass analyzer, that identifies with a high mass resolution (m/Δm reaching 10,000) the secondary ions, which are specific ion fragments of particular molecules, e.g. lipids, or particular molecular elements, e.g. protein amino acids or DNA nucleobases. In addition to spectroscopic mode, enabling micro-analysis of molecular composition, chemical imaging mode operates with a high lateral resolution (<100 nm). Surface sensitivity is characterized by exponential decay of signal intensity with (sub)nanometer constant. At the same time the signal can be acquired from whole protein monolayers.
To enhance detection of subtle differences in surface chemistry between different analysed samples, Principal Component Analysis is applied on a big set of multivariate ToF-SIMS data, where the recorded different spectra are visualised as points in multi-dimensional space with a coordinate system defined by various secondary ions signals. PCA determines the directions of the greatest uncorrelated variations within the data set, so called principal components, which form a new coordinate system. Also, PCA reduces dimensionality of the data set while retaining its essential information by transforming the original signals variables into only two (or three) principal components (e.g. PC1 and PC2). Finally, the resulting Scores plot describes surface compositions in terms of principal components, while the Loadings plot defines original signals that contribute to each principal component.
Recent applications of multivariate PCA analysis of ToF-SIMS data to control orientation, conformation and biorecognition of proteins on different surfaces, including self-assembled monolayers, conjugated polymers and thermo-responsive polymer brushes, are highlighted in our recent review1.
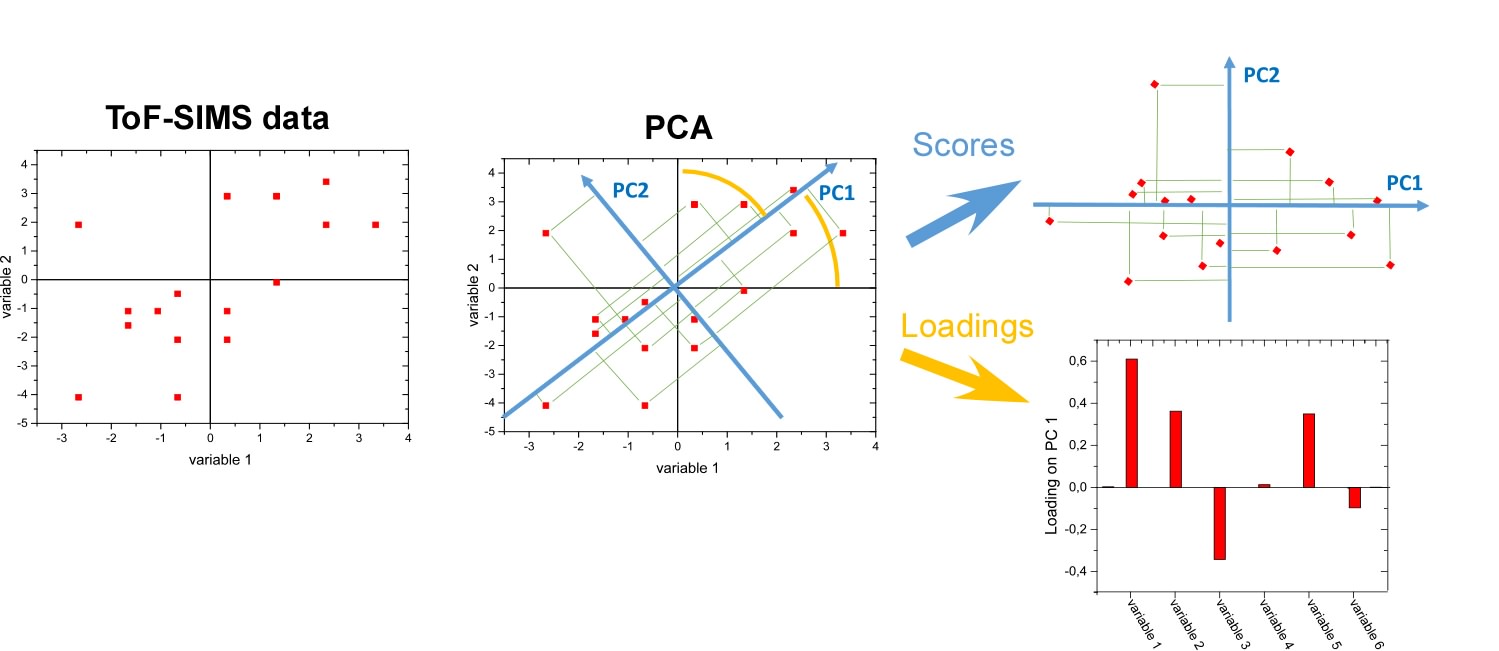
Fig.1. Principles of PCA analysis of ToF-SIMS data.
Over the past decade we have developed and applied several ToF-SIMS surface bioanalysis methodologies suitable for different research problems. Some of these methodologies are illustrated below.
Step-by-step characterization of biomolecular composition
ToF-SIMS can characterize and compare composition of multi-molecular layers resulting from subsequent surface modifications, e.g. due to multi-step functionalization, immobilization and/or assay. We analyze biomolecular multi-component layers consisting of a few different proteins (e.g. biosensors surfaces)2,3, or various proteins but also other biomolecules, such as DNA probes4,5 or lipids3 [originating from milk or phospholipids], or proteins and polymers6,7.
The simplest step-by-step characterization of biomolecular composition can be performed using characteristic ion signal intensities. For instance, the streptavidin-biotin system has been studied to immobilize biotinylated oligonucleotide probes onto surfaces functionalized with silane self-assembled monolayers. Chemical selectivity is provided by the signals characteristic for each of the biomolecules (BSA, Streptavidin, DNA) employed in the examined immobilization approaches4.
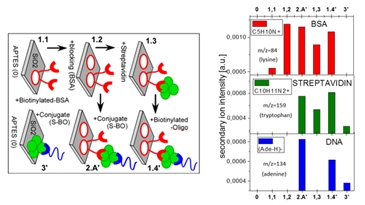
Fig.2. Step-by-step characterization of surface modifications with different biomolecules, based on characteristic ToF-SIMS signals.
More accurate step-by-step characterization and comparison of the multi-molecular layers composition is provided by multivariate PCA analysis. For instance, surface of on-chip biosensors has been examined in-situ after subsequent steps of the protocol for competitive immunodetection of κ-casein in goat milk. Surface composition is expressed (see the scores plot) in terms of principal components (here PC1 and PC3) that clearly distinguish between different biomolecules (milk glycerides from proteins, κ-casein protein from antibody)3.
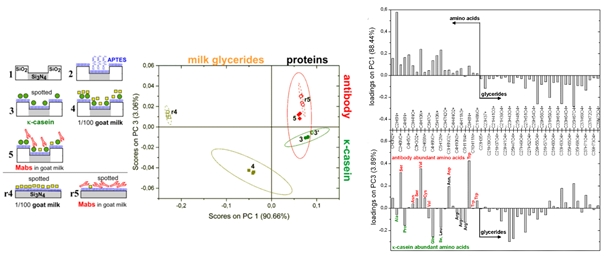
Fig.3. Step-by-step characterization of surface modifications with different biomolecules, based on PCA analysis of ToF-SIMS data.
Multivariate PCA characterization of orientation and conformation (denaturation) of proteins on surfaces
In the last decade, ToF-SIMS has been demonstrated as a powerful method to characterize changes in the conformation (denaturation) or orientation7,8 of biomolecules. Such molecular characterization benefits from interrogation of the information-rich ToF-SIMS data with multivariate Principal Component Analysis.
A characteristic example is the examination of temperature-controlled orientation of BSA protein, with resolved 3 domains (Albumin 1, 2, 3), on temperature-responsive grafted PBMA polymer brushes that exhibit morphological transformations around glass transition temperature Tg. Scores plot clearly shows that the principal component PC2 separates the data for BSA adsorbed on the brush below and above Tg (in contrast to BSA on a glass reference). Due to the extreme ToF-SIMS surface sensitivity, the resolved difference points to distinct amino acid composition of the outermost region of the adsorbed protein in these two temperature regions. The reason for this can be i) different protein conformation or ii) different protein orientation. To resolve this question two plots of the loadings (on PC2) are examined: Conformation (denaturation) changes, modifying the ratio of exposed hydrophilic and hydrophobic amino acids, can be excluded. This is because no systematic dependence can be noticed for the loadings (on PC2) plotted for each amino acid as a function of its side-chain hydrophobicity. In contrast, the negative values of the loadings (corresponding to T < Tg) can be related to the mass signals of amino acids more abundant in Albumin 1 and Albumin 2 than Albumin 3. In turn the principal component (PC2) is loaded positively (for T > Tg) by the fragments of amino acids rich in Albumin 3 when compared with Albumin 1 and Albumin 2. Therefore, changes in protein orientation can be concluded9.
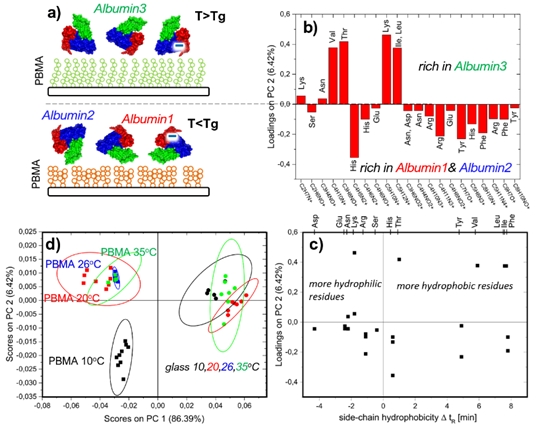
Fig.4. ToF-SIMS surface sensitivity to the outermost regions of immobilized proteins, enhanced by multivariate PCA analysis, enables examination of the protein state: Temperature-controlled orientation of protein with preserved conformation for BSA on temperature-responsive PBMA grafted brushes.
Molecular imaging of biomolecular patterns and cells
ToF-SIMS provides an unique opportunity of the chemical surface imaging with the label-free molecular discrimination and a sub-micrometer spatial resolution. We apply TOF-SIMS chemical imaging for evaluation of biomolecular patterns for biosensing and for analysis of the specific molecular markers distribution within cells.
For instance, ToF-SIMS chemical imaging of healthy and idiopathic pulmonary fibrosis derived lung fibroblasts has been applied to inspect differences in chemical properties of these cell lines. Maps of ions characteristic for cholesterol and phospholipids have revealed a significant difference in these species distribution between healthy and fibrotic cells10.

Fig.5. ToF-SIMS imaging of the cholesterol and phospholipids distribution within healthy (LL24) and fibrotic lung fibroblasts (LL97A).
Cancer cells differentiation based on whole spectra analysis (collaboration with prof. Małgorzata Lekka, IFJ PAN, Kraków, Poland)
The PCA has been applied to analyze the whole range of mass spectra without preselection of any particular masses and distinguish between various stages of cancer progression The PC3 versus PC2 plot has showed significant differences between nonmalignant cancer cells and the cancerous ones. Our findings show that the invasive phenotype of human bladder cancer cells can be correlated with the alterations in chemical compositions of the studied cells11,12.
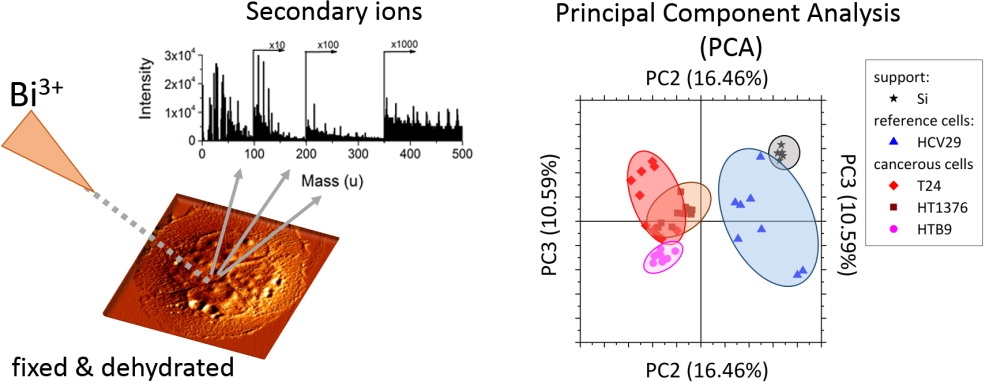
Fig.6. Application of TOF-SIMS with PCA multivariate data analysis for differentiation of cancerous cells.
Our research is performed in the frame of different projects:
Impact of polymer tacticity on adsorption, conformation and orientation of proteins on surface of thin polymer films (NCN UMO-2016/21/D/ST5/01633)
Multicomponent polymer films towards fabrication of multiprotein patterns (NCN DEC-2013/09/D/ST5/03860)
Protein immobilisation to film surfaces and micro-patterns of conjugated polymer in the aspect of biosensors applications (NCN UMO-2011/03/N/ST5/04764)
References:
1. "Controlling orientation, conformation, and biorecognition of proteins on silane monolayers, conjugate polymers, and thermo-responsive polymer brushes: investigations using TOF-SIMS and principal component analysis" K. Gajos, K. Awsiuk, A. Budkowski, Colloid and Polymer Science (2020), in press
2. "Indirect immunoassay on functionalized silicon surface: Molecular arrangement, composition and orientation examined step-by-step with multi-technique and multivariate analysis” K. Gajos, A. Budkowski, V. Pagkali, P. Petrou, M. Biernat, K. Awsiuk, J. Rysz, A. Bernasik, K. Misiakos, I. Raptis, S. Kakabakos, Colloids and Surfaces B: Biointerfaces 150 (2017), 437-444
3. "Imaging and chemical surface analysis of biomolecular functionalization of monolithically integrated on silicon Mach-Zehnder interferometric immunosensors" K. Gajos, M. Angelopoulou, P. Petrou, K. Awsiuk, S. Kakabakos, W. Haasnoot, A. Bernasik, J. Rysz, M. M. Marzec, K. Misiakos, I. Raptis, A. Budkowski, Applied Surface Science 385 (2016), 529-542
4. "Immobilization of oligonucleotide probes on silicon surfaces using biotin–streptavidin system examined with microscopic and spectroscopic techniques" K. Awsiuk, J. Rysz, P. Petrou, A. Budkowski, A. Bernasik, S. Kakabakos, M.M. Marzec, I. Raptis, Applied Surface Science 290 (2014) 199–206
5. "Imaging and spectroscopic comparison of multi-step methods to form DNA arrays based on the biotin-streptavidin system" K. Gajos, P. Petrou, A. Budkowski, K. Awsiuk, A. Bernasik, K. Misiakos, J. Rysz, I. Raptis, S. Kakabakos, Analyst 140 (2015), 1127–1139
6. "Sequential binary protein patterning on surface domains of thermo-responsive polymer blends cast by horizontal-dipping" J. Zemła, K. Gajos, K. Awsiuk, J. Rysz, A. Budkowski, Materials Science and Engineering C 99 (2019), 1477-1484
7. "Orientation and biorecognition of immunoglobulin adsorbed on spin-cast poly(3-alkylthiophenes): impact of polymer film crystallinity" K. Awsiuk, A. Budkowski, P. Petrou, M. M. Marzec, M. Biernat, T. Jaworska-Gołąb, J. Rysz, Colloids and Surfaces B: Biointerfaces 148 (2016), 278-286
8. "Orientation of Biotin-Binding Sites in Streptavidin Adsorbed onto the Surface of Polythiophene Films" K. Awsiuk, P. Petrou, A. Thanassoulas, J. Raczkowska, Langmuir 35 (2019), 3058-3066
9. "Temperature-Controlled Orientation of Proteins on Temperature-Responsive Grafted Polymer Brushes: Poly(butyl methacrylate) vs Poly(butyl acrylate): Morphology, Wetting, and Protein Adsorption" K. Awsiuk, Y. Stetsyshyn, J. Raczkowska, O. Lishchynskyi, P. Dabczyński, A. Kostruba, H. Ohar, Y. Shymborska, S. Nastyshyn, A. Budkowski, Biomacromolecules 20 (2019), 2185-2197
10. "Effect of Substrate Stiffness on Physicochemical Properties of Normal and Fibrotic Lung Fibroblasts" J. Raczkowska, B. Orzechowska, S. Patryas, K. Awsiuk, A. Kubiak, M. Kinoshita, M. Okamoto, J. Bobrowska, T. Stachura, J. Soja, K. Sładek, M. Lekka, Materials 13 (2020), 4495
11. "Biophysical and biochemical characteristics as complementary indicators of melanoma progression" J. Bobrowska, K. Awsiuk, J. Pabijan, P. Bobrowski, J. Lekki, K.M. Sowa, J. Rysz, A. Budkowski, M. Lekka, Analytical Chemistry 91 (2019), 9885-9892
12. "Comparing surface properties of melanoma cells using time of flight secondary ions mass spectrometry" J. Bobrowska, J. Moffat, K. Awsiuk, J. Pabijan, J. Rysz, A. Budkowski, M. Reading, M. Lekka, Analyst 141 (2016), 6217-6225
Contact:
Osoba publikująca: Kamil Awsiuk